-
Slides day 1
Exercise 1 - Parameter estimation
Exercise 2 - Tree topologies
Exercise 3 - Model comparison
Exercise 4 - Branch support
Exercise 5 - Command line
Exercise 6 - Inferring ML phylogenies with codon models
Exercise 7 - Inferring ML phylogenies using real datasets
Exercise 8 - Re-Analyze published datasets
Data formats in bioinformatics
Sequence and (Multi) Sequence Alignment formats
1. Fasta
In bioinformatics, FASTA format is a text-based format for representing either nucleotide sequences or peptide sequences, in which nucleotides or amino acids are represented using single-letter codes. The format also allows for sequence names and comments to precede the sequences.
>SEQUENCE_1
MTEITAAMVKELRESTGAGMMDCKNALSETNGDFDKAVQLLREKGLGKAAKKADRLAAEG
LVSVKVSDDFTIAAMRPSYLSYEDLDMTFVENEYKALVAELEKENEERRRLKDPNKPEHK
IPQFASRKQLSDAILKEAEEKIKEELKAQGKPEKIWDNIIPGKMNSFIADNSQLDSKLTL
MGQFYVMDDKKTVEQVIAEKEKEFGGKIKIVEFICFEVGEGLEKKTEDFAAEVAAQL
>SEQUENCE_2
SATVSEINSETDFVAKNDQFIALTKDTTAHIQSNSLQSVEELHSSTINGVKFEEYLKSQI
ATIGENLVVRRFATLKAGANGVVNGYIHTNGRVGVVIAAACDSAEVASKSRDLLRQICMH
2. Phylip
The PHYLIP file format stores a multiple sequence alignment. The format was originally defined and used in Joe Felsenstein’s PHYLIP package, and has since been supported by several other bioinformatics tools (e.g., RAxML).
The PHYLIP format comes in two different flavors: Interleaved and Sequential
-
Interleaved format
5 42 Turkey AAGCTNGGGC ATTTCAGGGT Salmo gair AAGCCTTGGC AGTGCAGGGT H. Sapiens ACCGGTTGGC CGTTCAGGGT Chimp AAACCCTTGC CGTTACGCTT Gorilla AAACCCTTGC CGGTACGCTT GAGCCCGGGC AATACAGGGT AT GAGCCGTGGC CGGGCACGGT AT ACAGGTTGGC CGTTCAGGGT AA AAACCGAGGC CGGGACACTC AT AAACCATTGC CGGTACGCTT AA -
Sequential format
5 42 Turkey AAGCTNGGGC ATTTCAGGGT GAGCCCGGGC AATACAGGGT AT Salmo gair AAGCCTTGGC AGTGCAGGGT GAGCCGTGGC CGGGCACGGT AT H. Sapiens ACCGGTTGGC CGTTCAGGGT ACAGGTTGGC CGTTCAGGGT AA Chimp AAACCCTTGC CGTTACGCTT AAACCGAGGC CGGGACACTC AT Gorilla AAACCCTTGC CGGTACGCTT AAACCATTGC CGGTACGCTT AA
Tree formats
1. Newick
The Newick tree format (or Newick notation or New Hampshire tree format) is a way of representing graph trees with edge lengths using parentheses and commas. It is a standard for representing trees in computer-readable form that makes use of the correspondence between trees and nested parentheses, noticed in 1857 by the famous English mathematician Arthur Cayley. If we have this rooted tree:
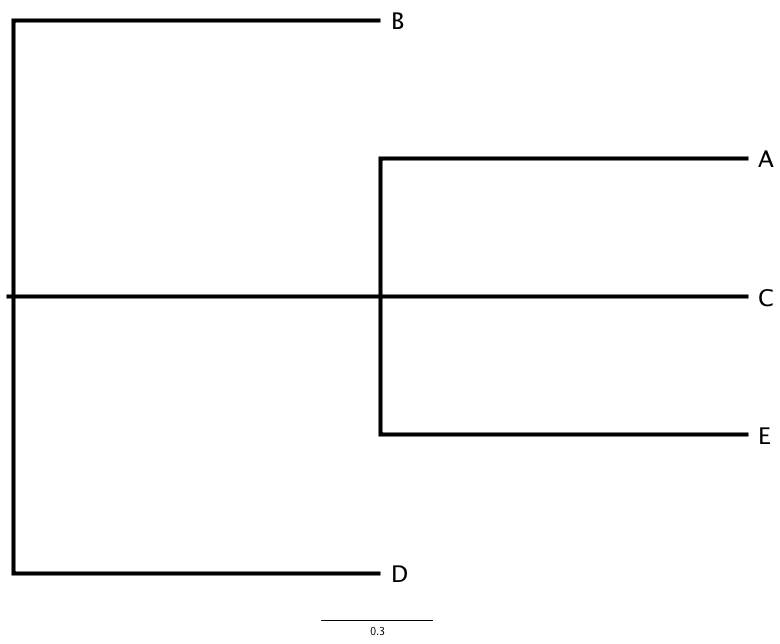
then in the tree file it is represented by the following sequence of printable characters:
(B,(A,C,E),D);
Branch lengths can be incorporated into a tree by putting a real number, with or without decimal point, after a node and preceded by a colon. This represents the length of the branch immediately below that node. Thus the above tree might have lengths represented as:
(B:6.0,(A:5.0,C:3.0,E:4.0):5.0,D:11.0);
2. PhyloXML
PhyloXML is an XML language for the analysis, exchange, and storage of phylogenetic trees (or networks) and associated data. The structure of phyloXML is described by XML Schema Definition (XSD) language.
<phyloxml xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:schemaLocation="http://www.phyloxml.org http://www.phyloxml.org/1.10/phyloxml.xsd"
xmlns="http://www.phyloxml.org">
<phylogeny rooted="true">
<name>example from Prof. Joe Felsenstein's book "Inferring Phylogenies"</name>
<description>MrBayes based on MAFFT alignment</description>
<clade>
<clade branch_length="0.06">
<confidence type="probability">0.88</confidence>
<clade branch_length="0.102">
<name>A</name>
</clade>
<clade branch_length="0.23">
<name>B</name>
</clade>
</clade>
<clade branch_length="0.4">
<name>C</name>
</clade>
</clade>
</phylogeny>
</phyloxml>
Multi purpose file formats
1. Nexux
The NEXUS file format (usually .nex or .nxs) is widely used in bioinformatics. Several popular phylogenetic programs such as PAUP*, MrBayes, Mesquite, MacClade and SplitsTree use this format.
#NEXUS
Begin data;
Dimensions ntax=4 nchar=15;
Format datatype=dna missing=? gap=-;
Matrix
A atgctagctagctcg
B atgcta??tag-tag
C atgttagctag-tgg
D atgttagctag-tag
;
End;
Begin Taxa;
Taxalabels A B C D;
End;
Begin Trees;
Tree tree1 = ((A,B),(C,D));
End;
- https://en.wikipedia.org/wiki/FASTA_format
- http://scikit-bio.org/docs/0.2.3/generated/skbio.io.phylip.html
- http://evolution.genetics.washington.edu/phylip/newicktree.html
- https://en.wikipedia.org/wiki/Nexus_file
- https://en.wikipedia.org/wiki/PhyloXML