-
Slides day 1
Exercise 1 - Parameter estimation
Exercise 2 - Tree topologies
Exercise 3 - Model comparison
Exercise 4 - Branch support
Exercise 5 - Command line
Exercise 6 - Inferring ML phylogenies with codon models
Exercise 7 - Inferring ML phylogenies using real datasets
Exercise 8 - Re-Analyze published datasets
How to visualise phylogenetic trees and alignments
Visualising Multi-Sequence-Alignments using Aliview
Discovering nucleotide/amino-acid variations in a MSA can be tricky. One way to help the user to overcome this issue is using dedicated softwares that allow basic operations on the MSA object (i.e. alignment browsing, coloring, indexing and nucleotide/amino-acid translation).
1. Open a MSA file
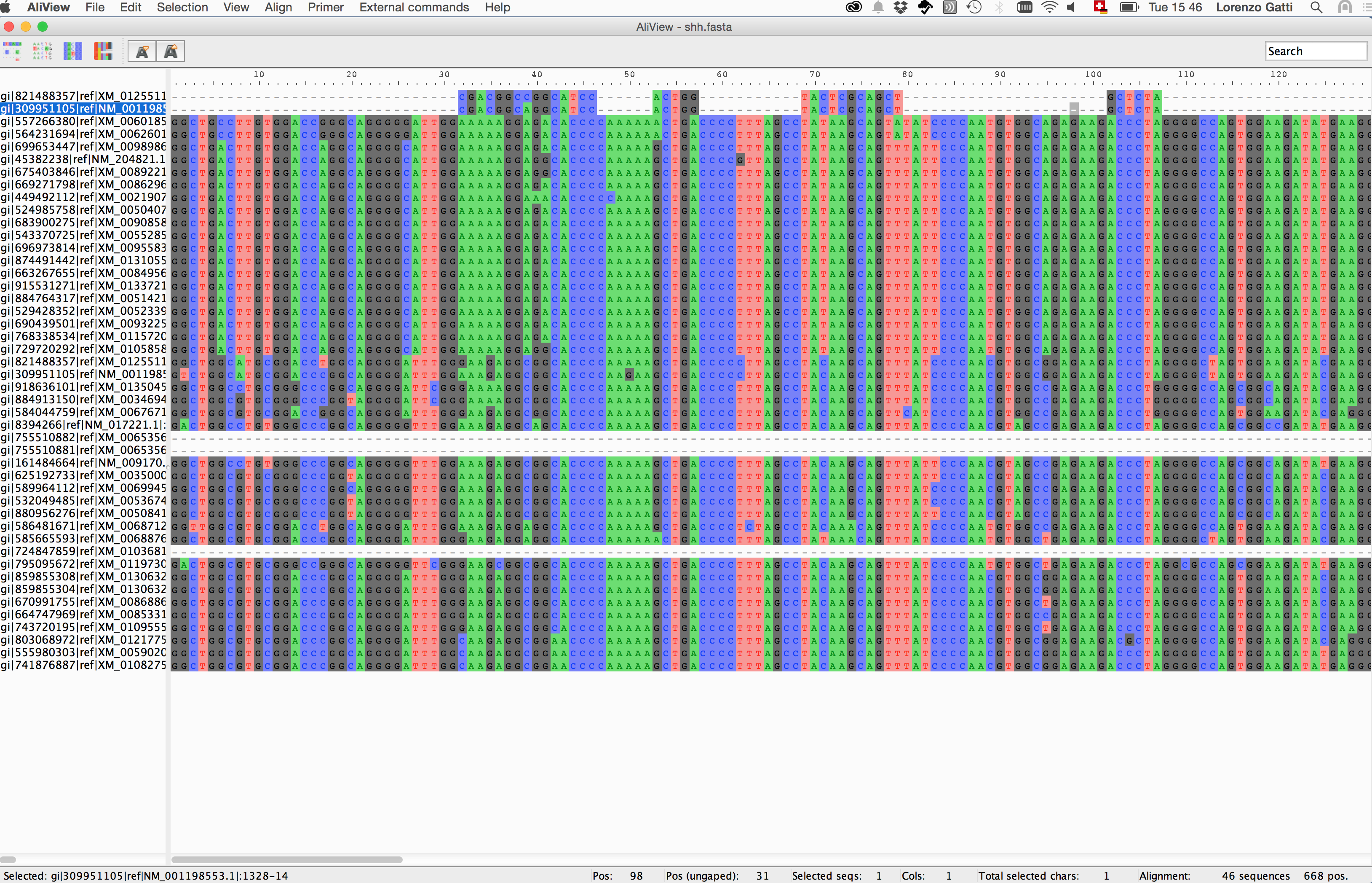
2. Perform a view
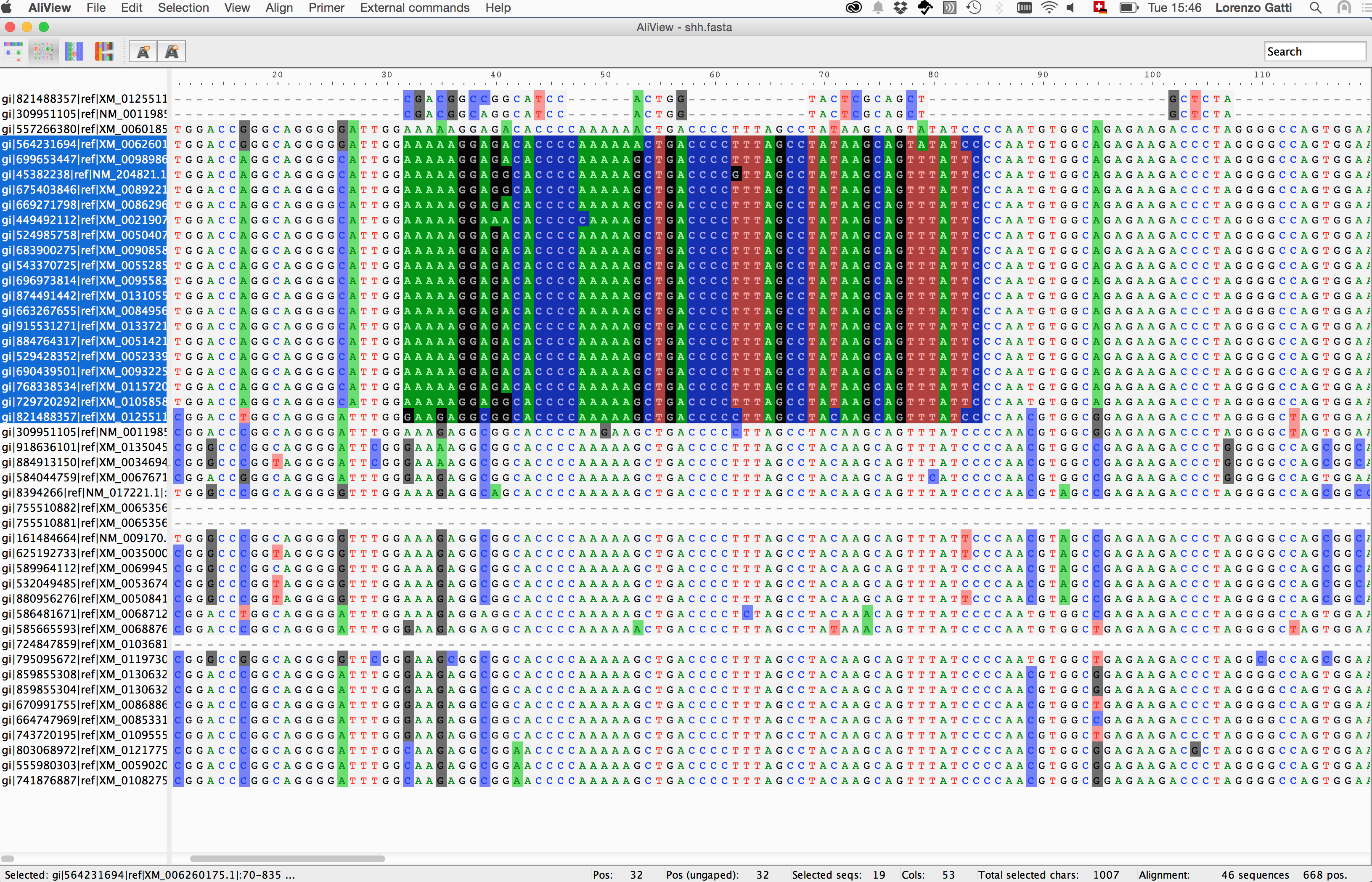
3. Extract view
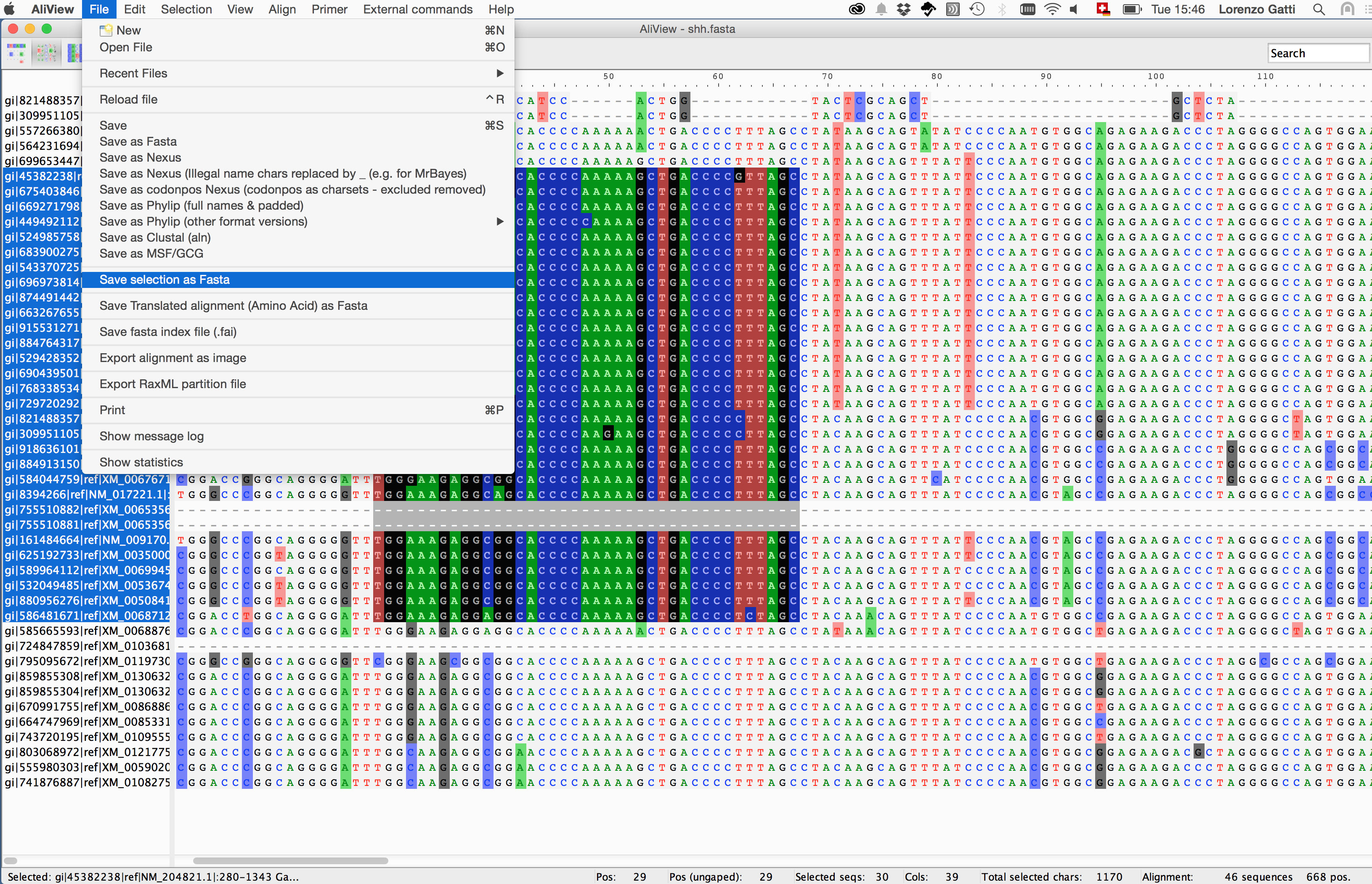
4. Summary statistics on the view

5. Translate a selection view using different codon frames
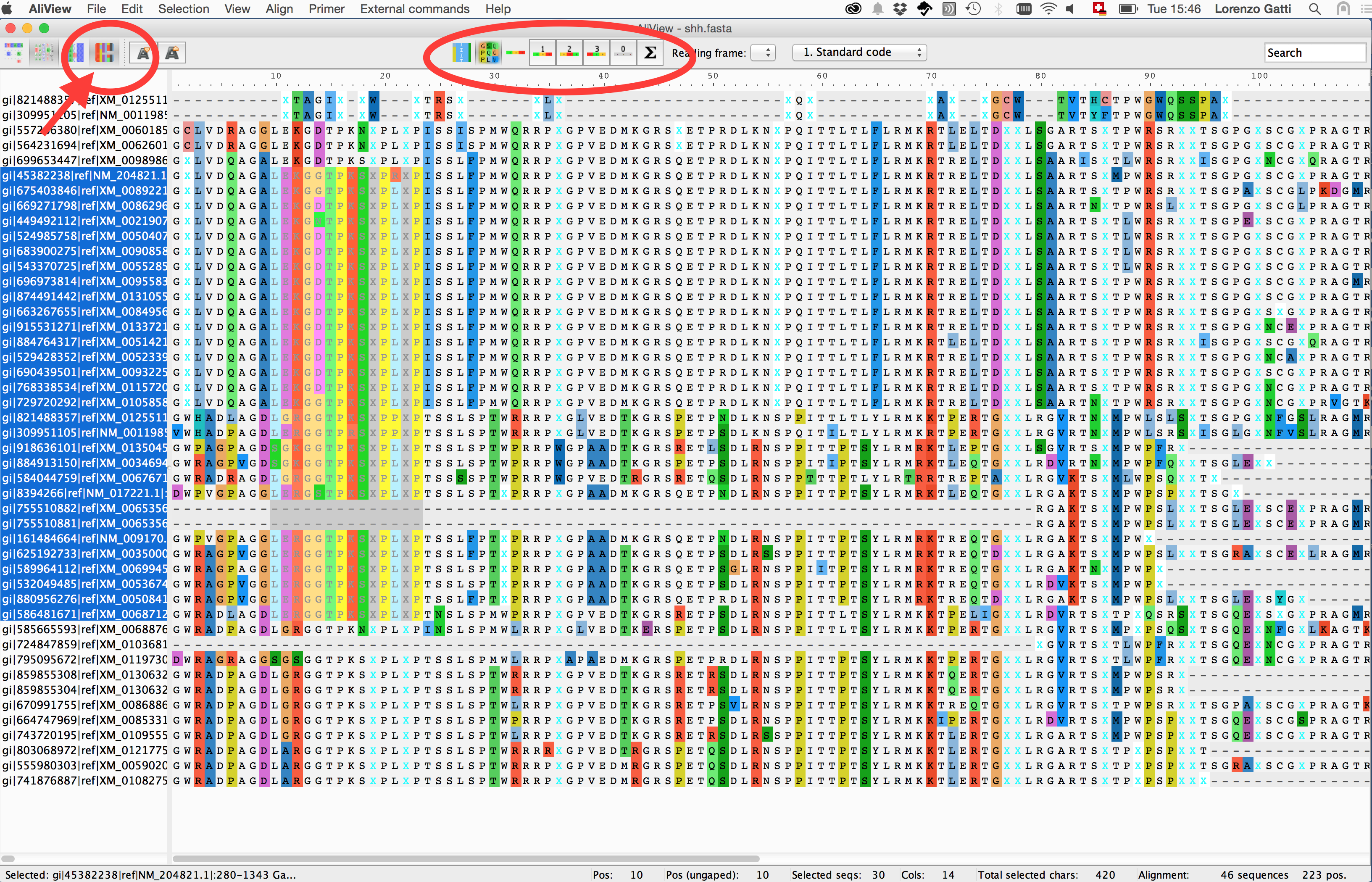
Other visualization softwares
You can find a list of MSA visualizers here: https://en.wikipedia.org/wiki/List_of_alignment_visualization_software
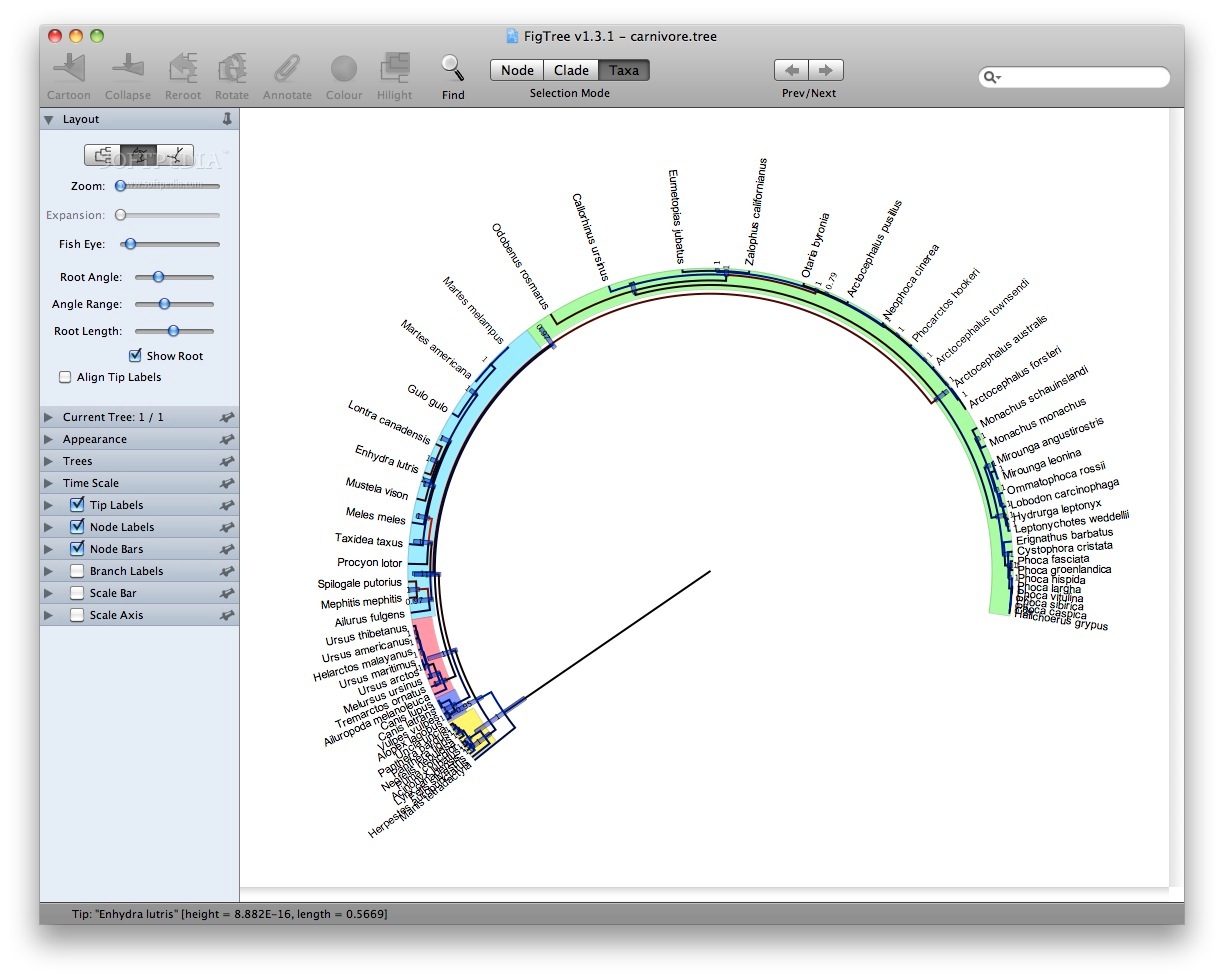
Visualizing phylogenetic trees with FigTree